Statistical
Machine Learning class, STAT-427/627, Dr. M. Baron
PRINCIPAL COMPONENTS AND PARTIAL LEAST SQUARES
1.
Eigenvalues and eigenvectors. Spectral decomposition.
>
attach(Auto);
>
library(ISLR2); attach(Auto);
>
X = model.matrix( mpg ~ weight + horsepower +
cylinders - 1, data=Auto )
>
A = cor(X)
>
A
weight
horsepower cylinders
weight 1.0000000 0.8645377
0.8975273
horsepower
0.8645377 1.0000000 0.8429834
cylinders 0.8975273 0.8429834
1.0000000
>
> eigen(A) #
This produces eigenvalues and eigenvectors
eigen()
decomposition
$values
[1]
2.73689293 0.16323918 0.09986789
$vectors
[,1] [,2] [,3]
[1,]
-0.5829268 0.2531535 0.7720813
[2,]
-0.5708026 -0.8038435 -0.1673921
[3,]
-0.5782566 0.5382834 -0.6130826
>
lambda = eigen(A)$values
>
Q = eigen(A)$vectors
>
> #
Check QQ' = Q'Q = I and the spectral decomposition
>
>
Q %*% t(Q)
[,1] [,2] [,3]
[1,]
1.000000e+00 5.551115e-17 2.220446e-16
[2,]
5.551115e-17 1.000000e+00 8.326673e-17
[3,]
2.220446e-16 8.326673e-17 1.000000e+00
>
t(Q) %*% Q
[,1] [,2] [,3]
[1,] 1.000000e+00
-5.551115e-17 -1.110223e-16
[2,]
-5.551115e-17 1.000000e+00 1.665335e-16
[3,]
-1.110223e-16 1.665335e-16 1.000000e+00
>
> #
These are identity matrices, all off-diagonal elements are practically 0
>
>
LAMBDA = diag(lambda) #
Diagonal matrix with eigenvalues on the diagonal
>
lambda
[1]
2.73689293 0.16323918 0.09986789
>
LAMBDA
[,1] [,2] [,3]
[1,]
2.736893 0.0000000 0.00000000
[2,]
0.000000 0.1632392 0.00000000
[3,]
0.000000 0.0000000 0.09986789
>
Q %*% LAMBDA %*% t(Q) #
Spectral decomposition of matrix A
[,1] [,2] [,3]
[1,]
1.0000000 0.8645377 0.8975273
[2,]
0.8645377 1.0000000 0.8429834
[3,]
0.8975273 0.8429834 1.0000000
>
A
weight
horsepower cylinders
weight 1.0000000 0.8645377
0.8975273
horsepower
0.8645377 1.0000000 0.8429834
cylinders 0.8975273 0.8429834
1.0000000
>
>
# This is the same matrix: Q %*% LAMBDA %*% t(Q) = A.
2.
Principal Components
# Let’s investigate the principal components, and how much variance
they explain.
> X = model.matrix( mpg
~ .-name-origin+as.factor(origin), data=Auto )
> pc = princomp(X)
> summary(pc)
Importance
of components:
Comp.1 Comp.2 Comp.3 Comp.4 Comp.5
Standard
deviation 854.5664182 38.860050688
1.614144e+01 3.309297e+00 1.694518e+00
Proportion
of Variance 0.9975617 0.002062789 3.559039e-04 1.495959e-05
3.922297e-06
Cumulative
Proportion 0.9975617 0.999624521 9.999804e-01 9.999954e-01
9.999993e-01
Comp.6 Comp.7 Comp.8 Comp.9
Standard
deviation 5.242357e-01 4.162175e-01
2.443204e-01 1.110223e-16
Proportion
of Variance 3.754062e-07 2.366403e-07 8.153944e-08 1.683715e-38
Cumulative
Proportion 9.999997e-01 9.999999e-01
1.000000e+00 1.000000e+00
# So, Z1, the first PC, contains 99.76% of the total
variation of X variables. The first two PCs together contain 99.96%. Here is a
plot of these percents called a screeplot.
> screeplot(pc)
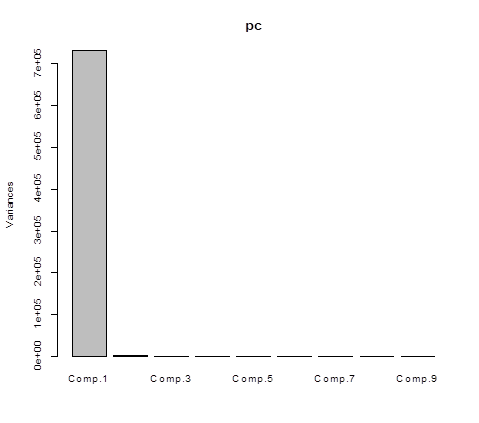
# The actual coefficients can be obtained by prcomp().
> prcomp(X)
PC1 PC2 PC3 PC4 PC5
(Intercept) 0.0000000000 0.0000000000
0.000000000 0.000000000 0.000000e+00
cylinders -0.0017926225 0.0133245279 -0.007294275 0.001414710
1.719368e-02
displacement -0.1143412856 0.9457785881 -0.303312504 -0.009143349
-1.059355e-02
horsepower -0.0389670412 0.2982553337
0.948761071 -0.043076559 -8.646402e-02
weight -0.9926735354 -0.1207516411
-0.002454212 0.001480458 3.152970e-03
acceleration 0.0013528348 -0.0348264293
-0.077006895 0.059516278 -9.944974e-01
year 0.0013368415 -0.0238516081
-0.042819254 -0.996935229 -5.549653e-02
as.factor(origin)2 0.0001308250 -0.0024889942 0.002857670
0.022100094 -9.052576e-05
as.factor(origin)3 0.0002103564 -0.0003765828 0.004796684 -0.012089823 -1.150938e-03
PC6 PC7 PC8 PC9
(Intercept) 0.0000000000 0.0000000000
0.000000e+00 1
cylinders 0.9911554803 0.1211162208 -4.909265e-02 0
displacement -0.0146594359 -0.0006512752 4.394368e-03
0
horsepower 0.0038232742 0.0034425206 -4.435100e-03 0
weight -0.0002093216 -0.0003053766 5.729471e-06
0
acceleration 0.0168319859 0.0012233398 -1.799780e-03 0
year -0.0001647840 0.0240346554
7.643176e-03 0
as.factor(origin)2
-0.0483462982 0.6888706846 7.229226e-01
0
as.factor(origin)3 0.1214929883 -0.7142804151 6.891098e-01
0
Standardized scale
So, we see that the 1st principal component contains a huge portion
of the total variation of X variables, and it is dominated by variable
“weight”. Of course! Looking at the
data, we see that weight simply has the largest values.
> head(Auto)
mpg cylinders displacement horsepower weight
acceleration year origin
1 18
8 307 130
3504 12.0 70
1
2 15
8 350 165
3693 11.5 70
1
3 18
8 318 150
3436 11.0 70
1
4 16
8 304 150
3433 12.0 70
1
5 17
8 302 140
3449 10.5 70
1
6 15
8 429 198
4341 10.0 70
1
# For this reason, usually, X variables are standardized first
(subtract each X-variable’s mean and divide by the standard deviation).
> pcr.fit = pcr( mpg ~ .-name-origin+as.factor(origin),
data=Auto, scale=TRUE
)
> summary(pcr.fit)
TRAINING:
% variance explained
1 comps 2 comps
3 comps 4 comps 5 comps
6 comps 7 comps 8 comps
X 56.9
73.02 84.29 92.38
97.29 98.86 99.59
100.00
mpg 71.8
73.64 73.96 79.25
79.25 80.22 81.55
82.42
Visualization
> names(USArrests)
[1]
"Murder"
"Assault" "UrbanPop" "Rape"
> pc = prcomp(USArrests, scale=TRUE)
> biplot(pc)
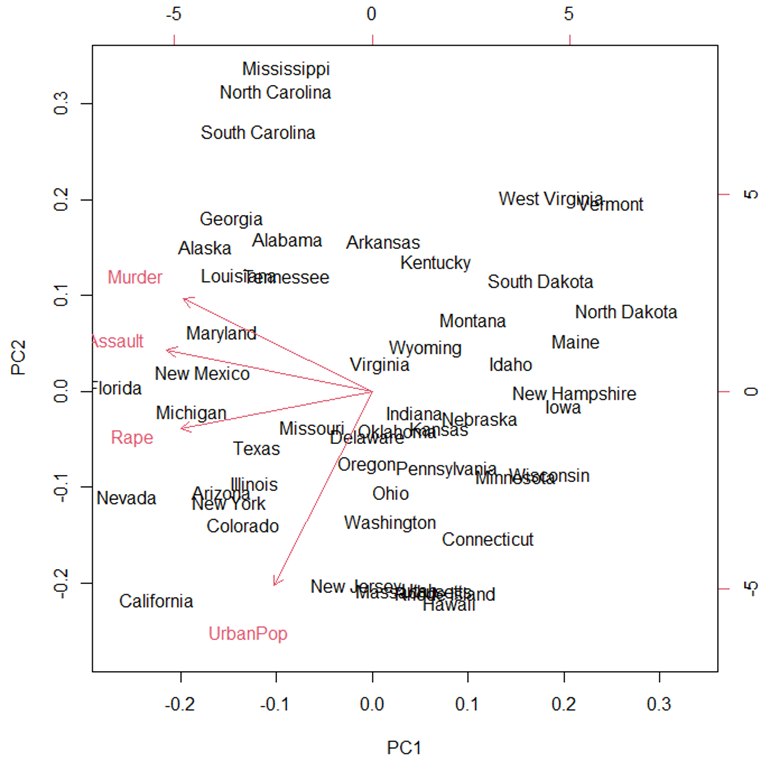
3.
Principal Components Regression
> library(pls)
> pcr.fit
= pcr(
mpg ~ . - name - origin + as.factor(origin), data=Auto )
# Using all variables except name
> summary(pcr.fit)
TRAINING:
% variance explained
1 comps 2 comps
3 comps 4 comps 5 comps
6 comps 7 comps 8 comps
X 99.76
99.96 100.00 100.00
100.00 100.00 100.00
100.00
mpg 69.35
70.09 70.75 80.79
80.88 80.91 80.93
82.42
# The “X” row shows % of X variation contained in the given number of
PCs.
# The “mpg” row shows R2 (% of Y variation explained) from
the PC regression. The usual linear regression on all 8 variables has the same
R2 as PCR that uses all 8 principal components.
> reg = lm( mpg ~
.-name-origin+as.factor(origin), data=Auto )
> summary(reg)
Multiple R-squared: 0.8242
Cross-validation
# Cross-validation. Option validation="CV" does a K-fold cross-validation with K=10, for LOOCV, use validation="LOO".
> pcr.fit = pcr( mpg ~ .-name-origin+as.factor(origin),
data=Auto, scale=TRUE, validation="CV"
)
> summary(pcr.fit)
VALIDATION:
RMSEP
Cross-validated
using 10 random segments.
(Intercept) 1 comps 2 comps
3 comps 4 comps 5 comps
6 comps 7 comps 8 comps
CV 7.815 4.162
4.036 4.028 3.611
3.616 3.537 3.427
3.350
adjCV
7.815 4.161 4.034
4.026 3.607 3.613
3.533 3.422 3.346
# The predicted error (by cross-validation) is minimized by using all
M=8 principal components.
# We can see the graph of root
mean-squared error of prediction (or specify val.type)
> validationplot(pcr.fit)
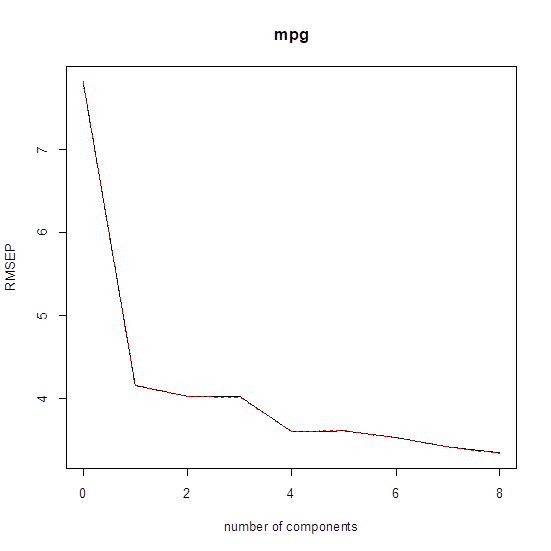
3.
Partial Least Squares
# Similar commands, just replace “pcr” with
“plsr”. M=6 components gives
the lowest prediction MSE.
> pls = plsr( mpg ~ .-name-origin+as.factor(origin), data=Auto, scale=TRUE,
validation="CV" )
> summary(pls)
Cross-validated using 10
random segments.
(Intercept) 1 comps 2 comps
3 comps 4 comps 5 comps
6 comps 7 comps 8 comps
CV 7.815 3.994
3.616 3.540 3.395
3.379 3.351 3.364
3.362
adjCV 7.815
3.992 3.612 3.535
3.390 3.376 3.345 3.359 3.357
TRAINING: % variance
explained
1 comps 2 comps
3 comps 4 comps 5 comps
6 comps 7 comps 8 comps
X 56.73
68.84 80.75 84.08
93.48 94.88 99.33
100.00
mpg 74.32
79.37 80.29 81.71
82.00 82.35 82.38
82.42
> validationplot(pls)
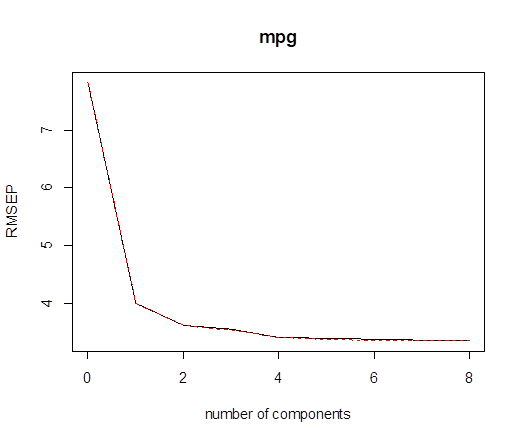
# Next, we can fit a model with the desired number of principal
components, obtain predicted values, and calculate the prediction mean-squared
error. For example:
> n = length(mpg);
> Z = sample(n,n/2);
> PLS = plsr( mpg ~ .-name-origin+as.factor(origin), data=Auto[Z,], scale=TRUE, ncomp=6 );
> Yhat = predict( PLS, newdata=Auto[-Z,], ncomp=6 )
> MSE = mean((Yhat -
mpg[-Z])^2)
> MSE
[1] 9.619134
> RMSE = sqrt(MSE)
> RMSE
[1] 3.101473